real_time_line_plotter.py source
This example performs image segmentaton on a stream of ultrasound images with a neural network, calculates the area of the segmentation regions and then plots the area curves over time using the LinePlotter.
This example performs image segmentaton on a stream of ultrasound images with a neural network, calculates the area of the segmentation regions and then plots the area curves over time using the LinePlotter.
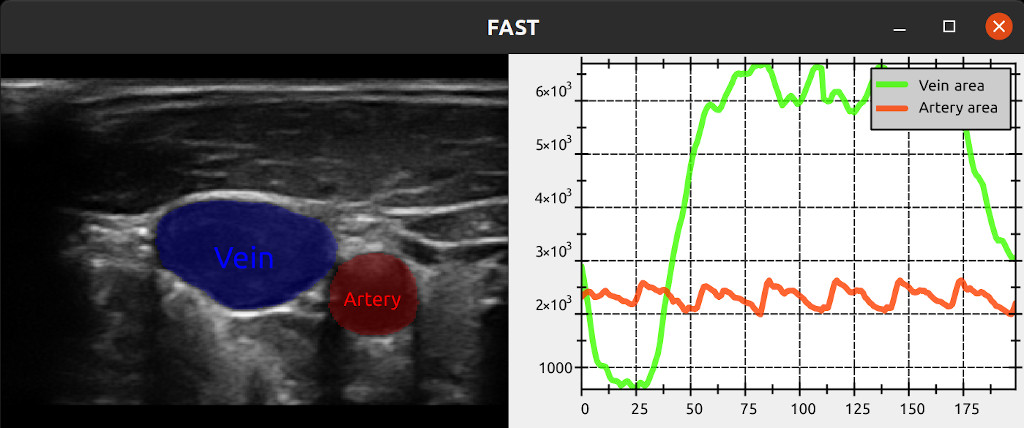
import platform if platform.system() != 'Windows': import PySide2.QtSvg # Must import this before fast due to conflicting symbols import fast # Must import FAST before rest of pyside2 from PySide2.QtWidgets import * from shiboken2 import wrapInstance import numpy as np fast.downloadTestDataIfNotExists() # This will download the test data needed to run the example class CalculateArea(fast.PythonProcessObject): """ Simple process object for calculating area of a two class segmentation """ def __init__(self): super().__init__() self.createInputPort(0) self.createOutputPort(0) self.createOutputPort(1) def execute(self): input = self.getInputData() input = np.asarray(input) area1 = float(np.sum(input == 1)) area2 = float(np.sum(input == 2)) self.addOutputData(0, fast.FloatScalar.create(area1)) self.addOutputData(1, fast.FloatScalar.create(area2)) # Set up computation pipeline streamer = fast.ImageFileStreamer.create( fast.Config.getTestDataPath() + 'US/JugularVein/US-2D_#.mhd', loop=True ) # Segmentation using a neural network segmentationNetwork = fast.SegmentationNetwork.create( fast.Config.getTestDataPath() + 'NeuralNetworkModels/jugular_vein_segmentation.xml', scaleFactor=1./255., ).connect(streamer) # Create area line plotting calculateArea = CalculateArea.create() \ .connect(segmentationNetwork) linePlotter = fast.LinePlotter.create(bufferSize=200,style=fast.PlottingStyle_DARK) \ .connect(calculateArea) \ .connect(1, calculateArea, 1) linePlotter.setNames({0: 'Artery area', 1: 'Vein area'}) # Visualization part imageRenderer = fast.ImageRenderer.create().connect(streamer) segmentationRenderer = fast.SegmentationRenderer.create( opacity=0.25, colors={1: fast.Color.Red(), 2: fast.Color.Blue()}, ).connect(segmentationNetwork) labelRenderer = fast.SegmentationLabelRenderer.create( labelNames={1: 'Artery', 2: 'Vein'}, labelColors={1: fast.Color.Red(), 2: fast.Color.Blue()}, ).connect(segmentationNetwork) # Setup window window = fast.SimpleWindow2D.create(bgcolor=fast.Color.Black(), width=1400, height=512)\ .connect([imageRenderer, segmentationRenderer, labelRenderer]) window.addProcessObject(linePlotter) # Use pyside2 to add plotting widget to the window mainWidget = wrapInstance(int(window.getWidget()), QWidget) layout = mainWidget.layout() plottingWidget = wrapInstance(int(linePlotter.getPlotterWidget()), QWidget) plottingWidget.setFixedWidth(700) plottingWidget.setFixedHeight(512) layout.addWidget(plottingWidget) # Run everything! window.run()