generate_tissue_patches_from_wsi.py source
This example loads a whole slide image (WSI), and generates a stream of patches of tissue in the WSI, and finally displays it using matplotlib.
This example loads a whole slide image (WSI), and generates a stream of patches of tissue in the WSI, and finally displays it using matplotlib
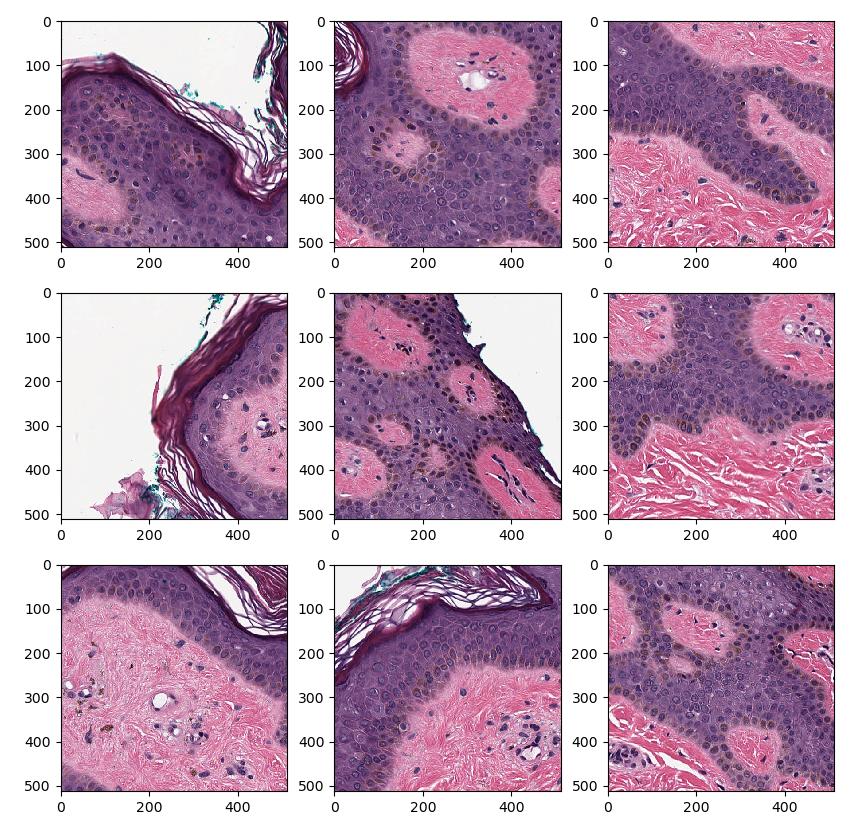
import fast import matplotlib.pyplot as plt # This will download the test data needed to run the example fast.downloadTestDataIfNotExists() importer = fast.WholeSlideImageImporter.create( fast.Config.getTestDataPath() + 'WSI/CMU-1.svs') tissueSegmentation = fast.TissueSegmentation.create().connect(importer) patchGenerator = fast.PatchGenerator.create(512, 512, level=0)\ .connect(0, importer)\ .connect(1, tissueSegmentation) # Create a 3x3 subplot for every set of 9 patches patch_list = [] for patch in fast.DataStream(patchGenerator): patch_list.append(patch) if len(patch_list) == 9: # Display the 9 last patches f, axes = plt.subplots(3,3, figsize=(10,10)) for i in range(3): for j in range(3): axes[i, j].imshow(patch_list[i + j*3]) plt.show() patch_list.clear() break